The Methylamp™ DNA Modification Kit is a complete set of optimized reagents designed to convert DNA so that 5-methylcytosines can be detected in downstream applications, for the purpose of gene-specific DNA methylation analysis. The kit has the following advantages and features: - Total protocol time is less than 2 hours.
- 99.9% conversion rate of unmethylated cytosine into uracil/thymine
- Extremely low degradation of DNA in the modification process, preventing over 90% of DNA loss.
- Requires as little as only 50 pg of input DNA, equivalent to about 20 cells.
- Extremely simple, robust, and reliable sodium bisulfite conversion protocol.
- Suitable for bisulfite sequencing (direct and cloning), next-generation sequencing, pyrosequencing, PCR (real-time, end-point, methylation-specific), COBRA (combined bisulfite restriction analysis), and methylation-based microarrays.
- Highly compatible with Illumina-based workflows.
Background Information
DNA methylation occurs by the covalent addition of a methyl group at the 5-carbon of the cytosine ring, resulting in 5-methylcytosine. DNA methylation is essential for the development of many species and is often associated with many key biological processes as well as gene regulation. There are various methods used to assess DNA methylation states. However, only bisulfite conversion of genomic DNA, followed by PCR amplification, cloning, and sequencing of individual PCR amplimers yields reliable information on the methylation states of individual cytosines on individual DNA molecules. By treating DNA with sodium bisulfite, cytosine residues are deaminated to uracil while leaving 5-methylcytosine intact, providing researchers with distinguishabe methylated positions in the DNA sequence during the downstream application. Principle & Procedure
DNA is chemically denatured to allow the sodium bisulfite reagent to react specifically with single-stranded DNA, thereby deaminating cytosine and creating a uracil residue. The unique DNA protection reagents contained in the modification buffer can prevent chemical and thermophilic degradation of DNA during the sodium bisulfite treatment process. The included non-toxic, modified DNA capture buffer enables the DNA to bind tightly to the column filter, permitting DNA cleaning to be carried out on the column to effectively remove residual sodium bisulfite and salts. Modified DNA can then be eluted for use in various methylation analysis applications or otherwise stored at -20°C for up to 2 months. 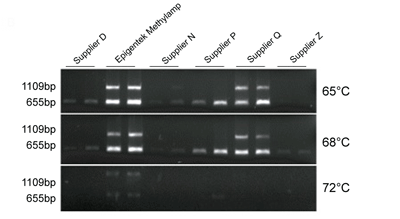
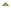 Fig. 3. Two bisulfite-converted DNAs from six kits, including the Methylamp DNA Modification Kit (Cat #P-1001), were subjected to PCR with two amplicons (655 and 1109 bp) and three different extension temperatures (65°C, 68°C and 72°C) to assess capacity for long amplicon amplification. Bisulfite-converted DNA from Epigentek's Methylamp kit with a PCR extension temperature of 65°C had the most robust amplification of the longer 1109 bp amplicon. Yang Y, et al. BMC Genomics. 2015; 16(1): 350. Fig. 3. Two bisulfite-converted DNAs from six kits, including the Methylamp DNA Modification Kit (Cat #P-1001), were subjected to PCR with two amplicons (655 and 1109 bp) and three different extension temperatures (65°C, 68°C and 72°C) to assess capacity for long amplicon amplification. Bisulfite-converted DNA from Epigentek's Methylamp kit with a PCR extension temperature of 65°C had the most robust amplification of the longer 1109 bp amplicon. Yang Y, et al. BMC Genomics. 2015; 16(1): 350.
| 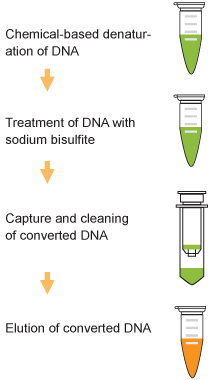
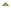 Fig. 1. Schematic procedure of the Methylamp™ DNA Modification Kit. Fig. 1. Schematic procedure of the Methylamp™ DNA Modification Kit.
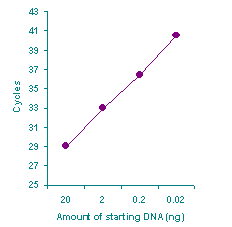
 Fig. 2. Different amounts of DNA isolated from a serum sample were chemically modified using the Methylamp™ DNA Modification Kit. Real time PCR was then performed by using a pair of primers and a probe designed to amplify both methylated and unmethylated Fig. 2. Different amounts of DNA isolated from a serum sample were chemically modified using the Methylamp™ DNA Modification Kit. Real time PCR was then performed by using a pair of primers and a probe designed to amplify both methylated and unmethylated
|
 HOME > 제품안내 > Epigenetics
HOME > 제품안내 > Epigenetics


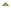 Fig. 3.
Fig. 3.
